Essential Tips for Selecting Mouse Models in Research
How do you decide which mouse model is most applicable for your research question or to test your therapeutic? With over 13,000 available mouse models at the Jackson Laboratory, it can be overwhelming and become a daunting task to decide. Whether you have a specific disease, gene, tissue, phenotype, or therapeutic in mind, there are four excellent resources to help you with model selection: JAX's Mouse Search, JAX's Strain Datasheets, the Mouse Genome Informatics (MGI), and the Mouse Phenome Database (MPD). This blog will walk you through how to maximize these JAX resources and use them together to inform your decision on which mouse model(s) might be best for your research.
In Brief:
- JAX's Mouse Search: User-friendly web interface designed to help researchers find specific mouse strains that suit their experimental needs.
- JAX's Strain Datasheets: These documents provide detailed information about specific mouse strains within JAX's repository, including how the model was created, its usage, phenotypic characteristics, breeding strategies, published references, legalities and more.
- Mouse Genome Informatics (MGI): This website offers comprehensive information about the genetics, genomics, biology, phenotype, and public availability of mouse strains harboring mutation(s) of interest. See “About MGI” for all the resources MGI offers.
- Mouse Phenome Database (MPD): A collaborative effort providing researchers with baseline phenotype and gene expression data, protocols, and SNP panels. The database includes anatomical, physiological, and behavioral data for a subset of mouse strains.
Where to Begin:
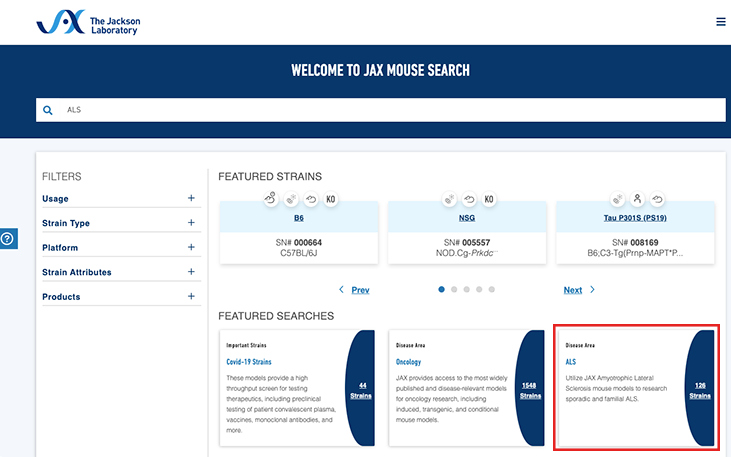
JAX's Mouse Search page and MGI are excellent starting points to explore publicly available mouse models in JAX's repository and all repositories, respectively. For MGI, searching by gene name is recommended. JAX's Mouse Search allows you to use keywords related to genotype, phenotype, or disease. JAX's Mouse Search page offers various filter settings to further refine the results based on “strain type”, “platform”, “strain attributes” and “product”. Additionally, curated lists of mouse models related to specific diseases and/or platforms can be found on JAX's Mouse Search homepage.
For example, if you are interested in amyotrophic lateral sclerosis (ALS) and unsure how to find related mouse models, go to the JAX Mouse Search page. There, you have multiple ways to initiate your search. One approach is to type “ALS” or your gene of interest into the search bar. Alternatively, you can explore the curated list of ALS strains listed under the “Featured Searches” section on the homepage. If you click on the “ALS Featured Search” the return gives over 100 strains related to ALS! Now that you have narrowed down your mouse model options to strains exhibiting the disease of interest, the question remains: how do you identify the most suitable disease model within this refined list to meet your specific research needs?
Further Refining Your Selection by Phenotype:
Exploring the genotype and phenotype of each strain can help further narrow down which mouse model(s) is most appropriate for your research question. JAX's Strain Datasheets serve as an excellent resource to understand each strain's genetic development and known phenotypes. The information on the strain datasheets is carefully curated from the published literature and communications with the donating investigator and/or strain creator. Additionally, in the “References” section of the strain datasheets you can find links to the original publication and other peer-reviewed articles.
Let's apply this to our ALS example. In our ALS example, the first two strains appear almost identical with matching descriptions on the search results, but you notice a difference in the strain name on the left side of the hyphen. As an aside, if you are interested in understanding mouse nomenclature, check out our on-demand webinar.
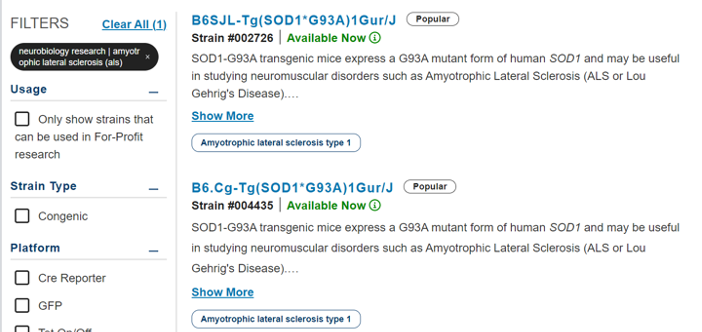
The mouse nomenclature links on your returned results will direct you to the related strain datasheet, where you can begin exploring the differences between the two models. In the “Development” section, which contains information about how the mouse was created, you discover that the primary distinction between the two strains lies in their genetic background (B6 in 004435 and B6SJL in 02726).
Delving into the strain datasheet “Detailed Description” section, which outlines phenotypic characteristics, you learn that due to the genetic background difference, the two models exhibit markedly different phenotypes. Mice on the B6J background (004435) have a 50% survival rate to 157.1+/-9.3 days, while those on the mixed B6SJL background where 50% survival is observed at 128.9+/-9.1 days.
Furthermore, the “Phenotype” section, which contains curated information from peer-reviewed publications, indicates mice on the mixed B6SJL background display severe muscle weakness beyond 3 months of age.
Further Refining Your Selection by Genotype:
To delve further into the genetics of the mutation(s), you can refer to the “Genetics” section of the Strain Datasheet, which includes all the alleles contributing to the mouse model.
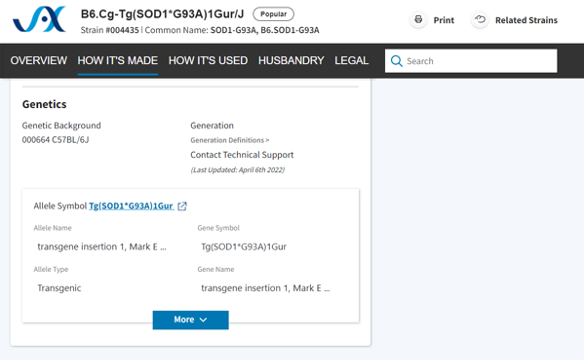
In our example, there is only one allele, Tg(SOD1*G93A)1Gur”. These alleles are linked to the Mouse Genome Informatics (MGI) Database. MGI can provide additional phenotype information, including details about the mutation's interactions with other genetic modifications and its effects on different genetic backgrounds. Moreover, for specific strains, both MGI and JAX Characterization Cre Lines provide data about gene expression.
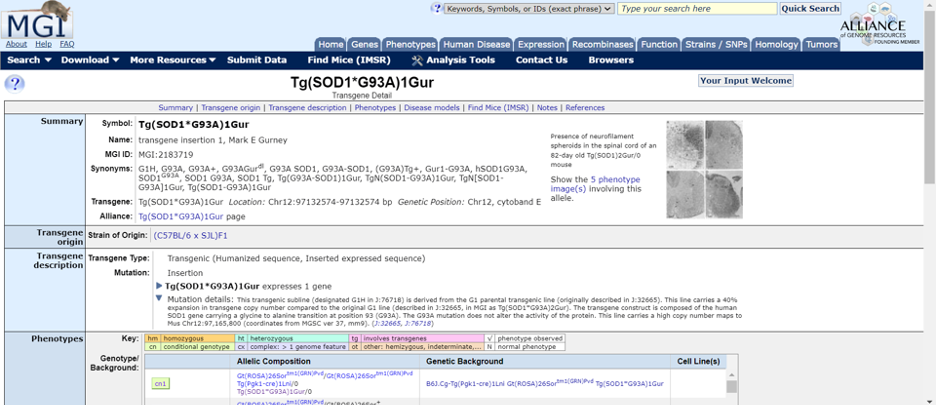
Clicking on the link in the strain datasheet for the Tg(SOD1*G93A)1Gur allele will redirect to the “Transgene Detail” page for the Tg(SOD1*G93A)1Gur allele on MGI. On this page, under the “Transgene origin” row, you can find which inbred strain the mutation was originally created in. (you can also find this information in the “Detailed Description” and “Genetics” sections on the JAX strain datasheet). In this case the mutation was created in a F1 hybrid of C57BL6 x SJL.
Further down on the page, you'll find a table labeled “Affected Systems” in the “Phenotypes” row. This table serves as a comprehensive resource, consolidating the published phenotypes associated with this allele in one place! By clicking on the check marks in the table, you view the phenotypes linked to this allele. References are listed under every check mark for further exploration.
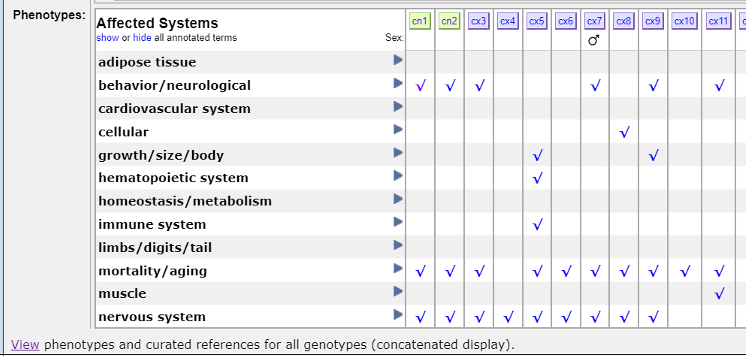
Each model is listed across the top- cn1, cn2, etc. By clicking on those labels, you can discover more about the phenotypes associated with each genotype.
Scrolling down the page under “Find Mice” you will find a list of publicly available mouse models that carry the desired allele. In this example, there are 4 strains available.
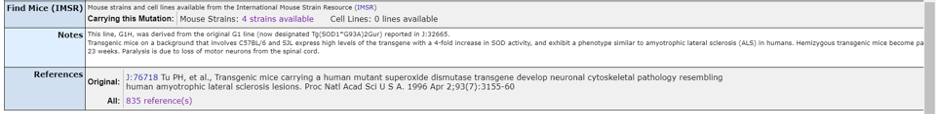
Additionally, the original reference for the allele is listed at the bottom of the page, along with all the curated references that mention using this specific allele. Remarkably, there are over 800 published papers utilizing this allele.
Present and Future Experimental Planning:
The Mouse Phenome Database (MPD) is another valuable tool for strain selection and experiment planning. This database integrates genomic and phenomic data, offering scientists access to actual datasets and protocols. The datasets in MPD encompass various strain types including: inbred, recombinant inbred, Collaborative Cross, chromosome substitution, mutants, and others. While not all strains found on the JAX website or in MGI will have datasets, the basic phenotypic characteristics of inbred strains will generally apply to most other mutant strains maintained on those backgrounds.
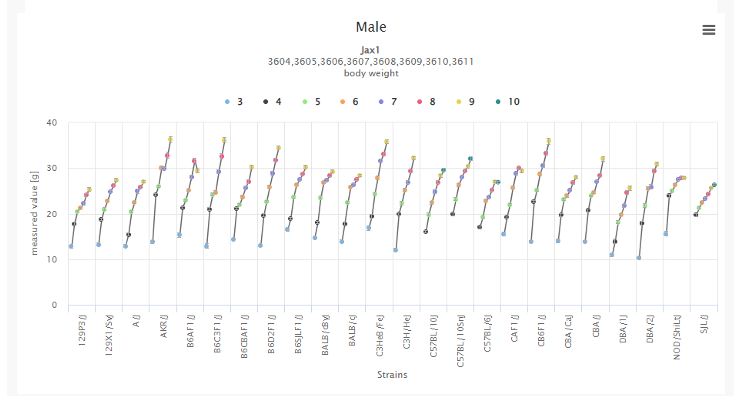
In the search function, you have the flexibility to search by JAX strain number, gene, assay type, data set name, and more. Datasets covering various phenotypes such as body weight, blood pressure, locomotor activity, blood glucose levels, and much more are available. A valuable application of the data in MPD is to consider whether an existing allele on a different genetic background might be more suitable for your research needs or to determine the most appropriate genetic background for generating a new allele.
Designing Your Own Mouse Models:
Finally, if the model you desire does not exist after going through the entire search process, don't worry! JAX offers model generation and breeding services, allowing us to create a custom, brand-new model for you or breed existing strains to create complex new combinations of alleles. JAX can use a variety of technologies such as CRISPR/CAS9 gene editing, DNA microinjection, BXB1 transgene insertion, and ES cell microinjection to generate your specific mouse model of interest.
For more information, see: Model Generation Services
Still have questions? Contact our Technical Information Services group at micetech@jax.org