The Mouse Tumor Biology (MTB) Database: Your First Stop in Finding a Tumor with Your Target of Interest
Searching for data
JAX provides researchers free access to the Mouse Tumor Biology (MTB) database, a continuously updated repository of tumor biology data. MTB supports the use of mice as a model system of human cancer and aids researchers in areas such as choosing experimental models, reviewing patterns of mutations in specific cancers, and identifying genes that are commonly mutated across a spectrum of cancers. There is a wealth of information at one’s fingertip, such as tumor frequency and kinetics data, genomic data, histology data, and standard of care (SOC) data.
So, how can you use the MTB database to meet your research needs?
Ready? Get Set… Search!
There are many ways to search the MTB database, including the Search Box; the search forms labeled Tumor, Strain, Genetics, Pathology Images, References and Advanced; Cancer QTL Viewer; or the Tumor Frequency Grid. Note that most of the search tools in the left side navigation will provide you with mouse tumor models. If you are specifically interested in human tumors, such as patient derived xenograft (PDX) heterologous tumors, you will need to select either “PDX Finder” or “PDX Model Search”.
In order to help you see how this works, we’ll run through an example search for a human PDX available from the library of tumors characterized at JAX. For example, if your research requires human breast cancer, specifically a triple-negative, invasive ductal carcinoma, you could find a PDX model tagged as being triple-negative, as well models that have tumor growth graphs and dosing studies. To find this specific model using the MTB database, first select “PDX Model Search” from the left-hand side navigation.
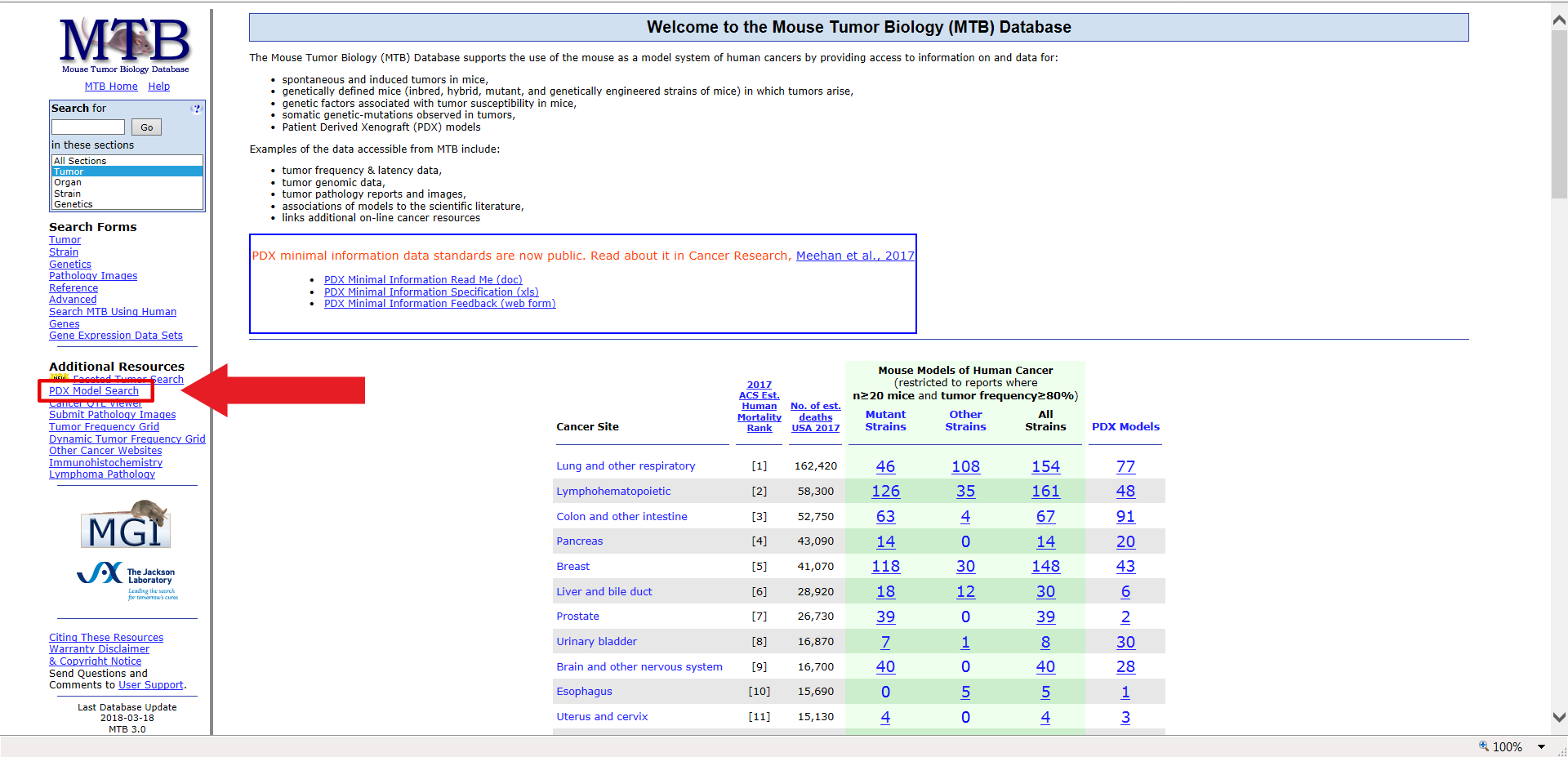
Under the selection criteria, select the criteria previously mentioned, and then limit the results to those that have both dosing study data and tumor growth graphs, and are tagged as “Triple Negative Breast Cancer (TBNBC)”. Under “Search by Diagnosis”, highlight “invasive ductal carcinoma”.
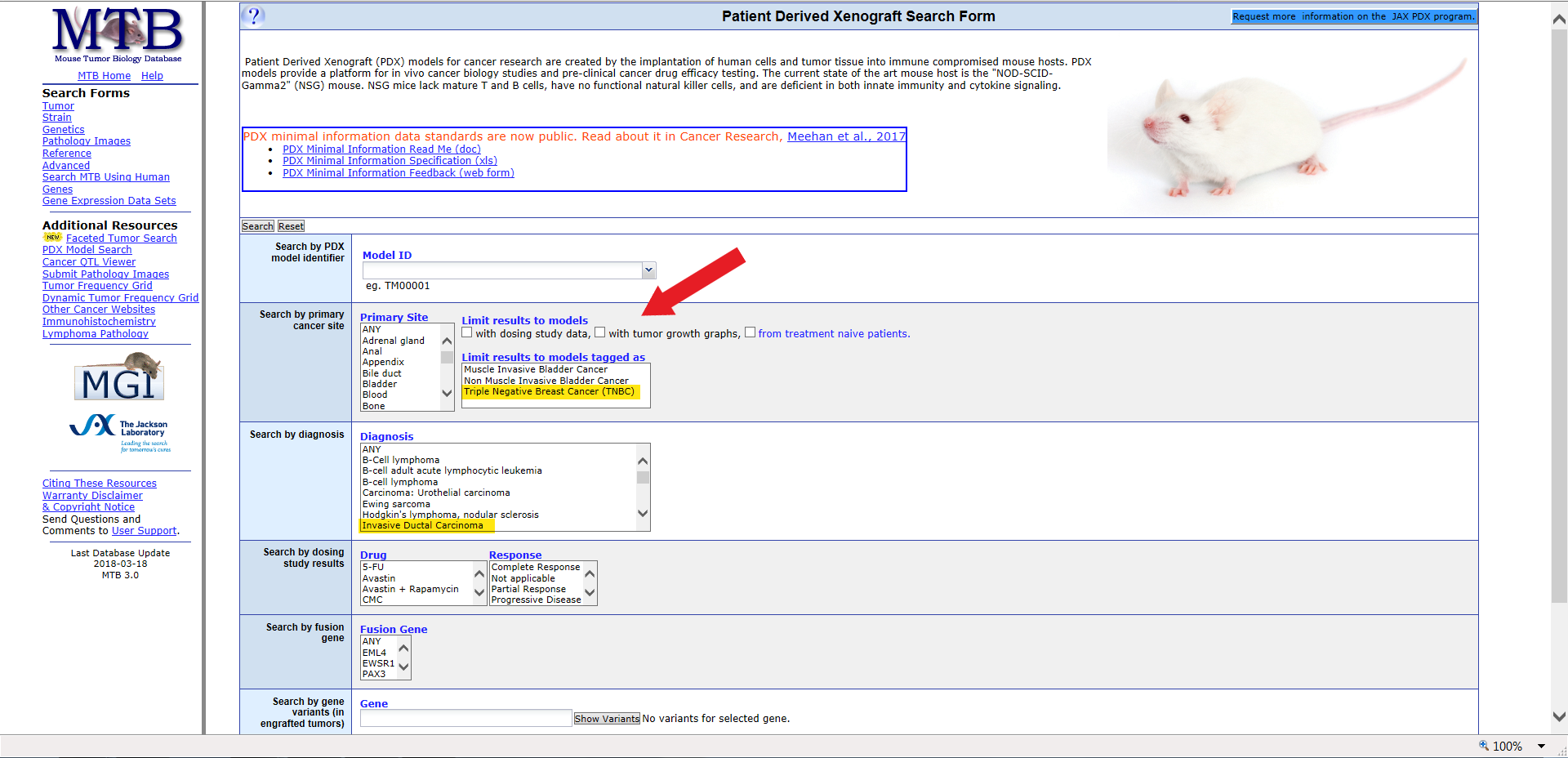
In this example, the search results in 2 possible matches. The result page also displays the primary site, diagnosis (initial: final), tumor site, tumor type, sex, age, and additional data.
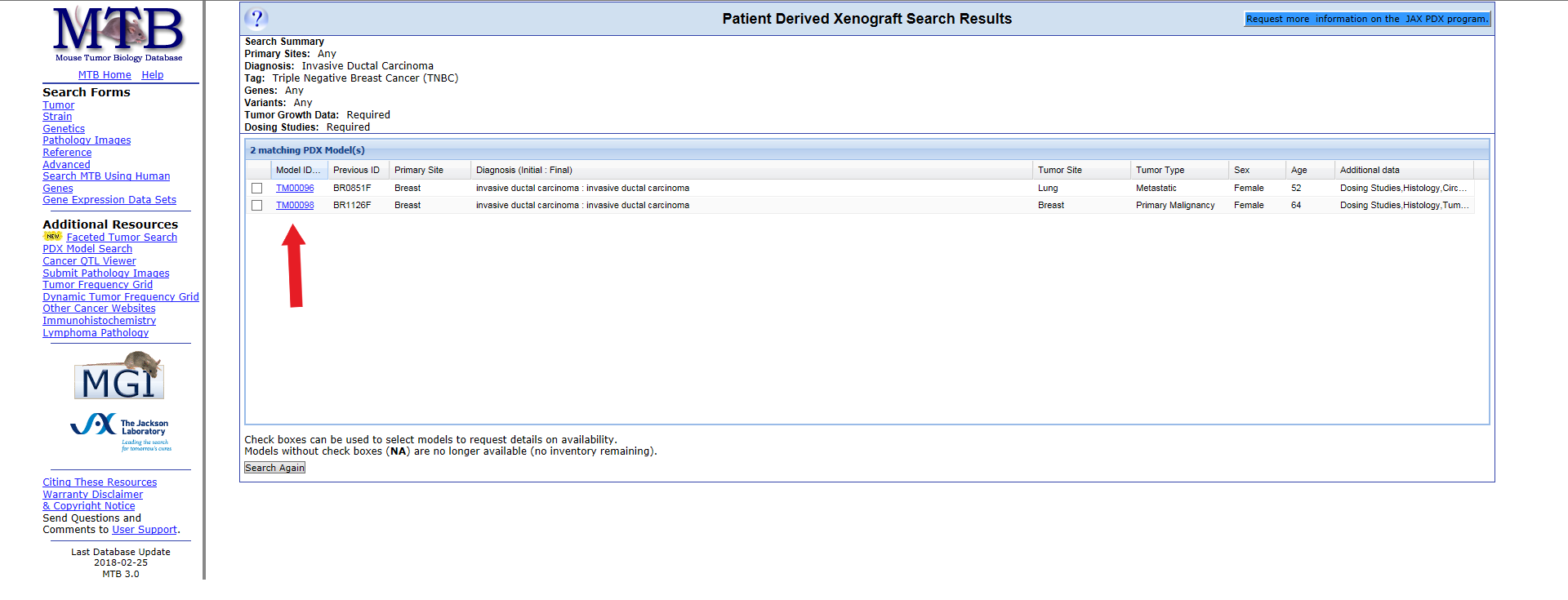
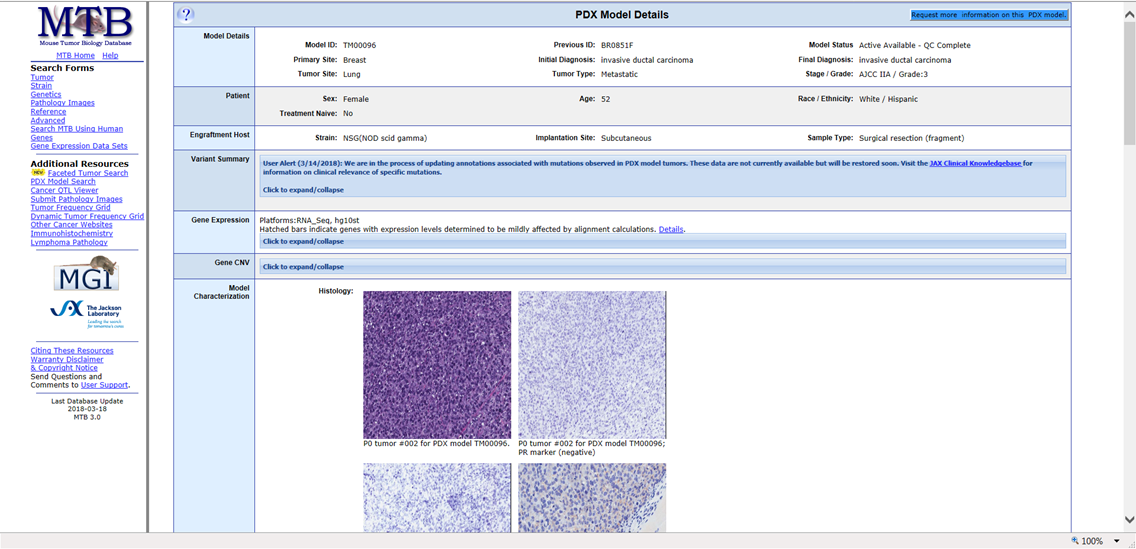
Click Model ID to open a full model detail page. This page lists details about the specific model including the ID, model status, stage or grade, tumor type, initial and final diagnoses, the engraftment host for the model, gene expression, histology images, tumor growth graphs, and dosing studies.
Let’s consider another example. Using the Patient Derived Xenograft Search Form within MTB, we can refine the PDX model search to those with a specific gene variant. Let’s search for gene variants for the BRCA1 gene, which has been shown to produce a protein involved in tumor suppression.
Starting from the MTB database main page, select the PDX Model Search from the left-hand side navigation. We will refine our search to include only triple-negative breast cancer models and search for the BRCA1 variants.
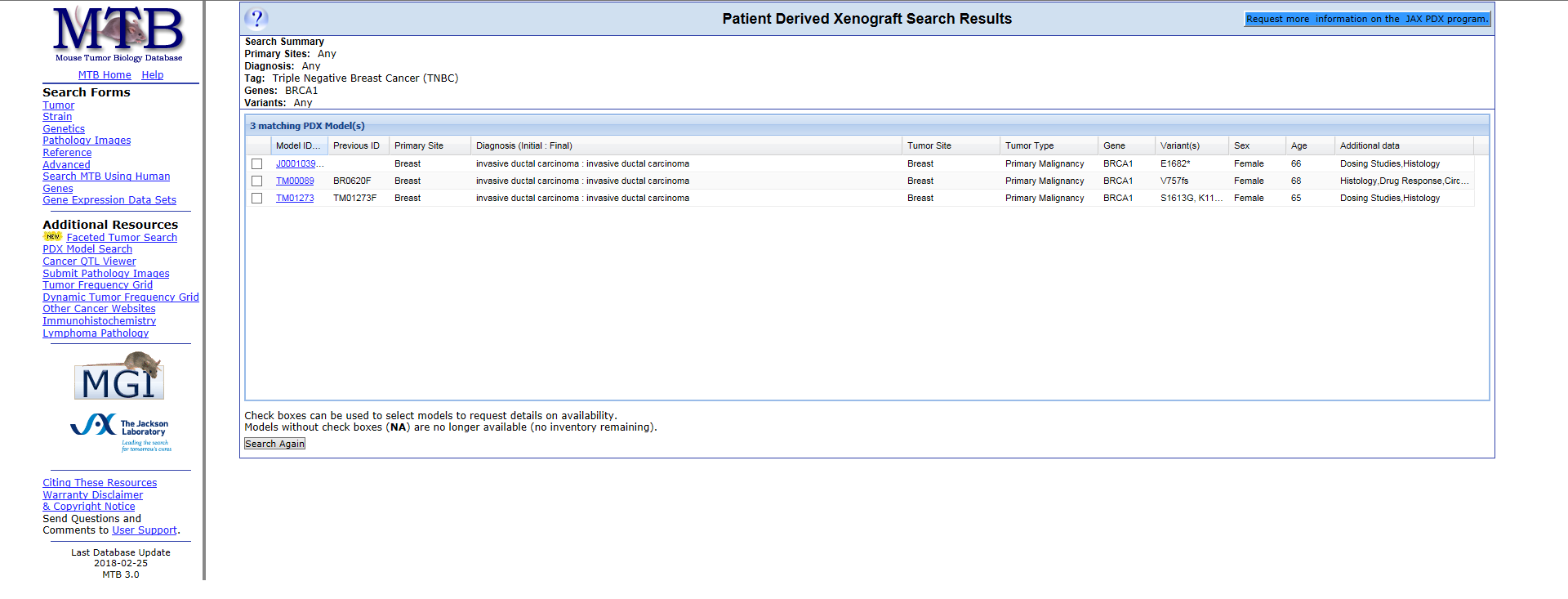
As with our previous example, the database searches for the closest matches based on the search criteria. This search yielded 3 possible triple-negative breast cancer models with variants annotated within the BRCA1 gene. The result page displays the models and details including the model ID, diagnosis (initial and final), tumor site and type, gene, variant(s), sex, age of the patient, and any additional details. For additional details on a specific model, click on the model ID.
MTB also provides gene expression data. For this example, let’s search for breast tumor models based on EGFR gene expression. Starting from the PDX Model Search page, select “breast” under “Search by primary cancer site”, and then enter “EGFR” under “Display a chart of gene expression”.
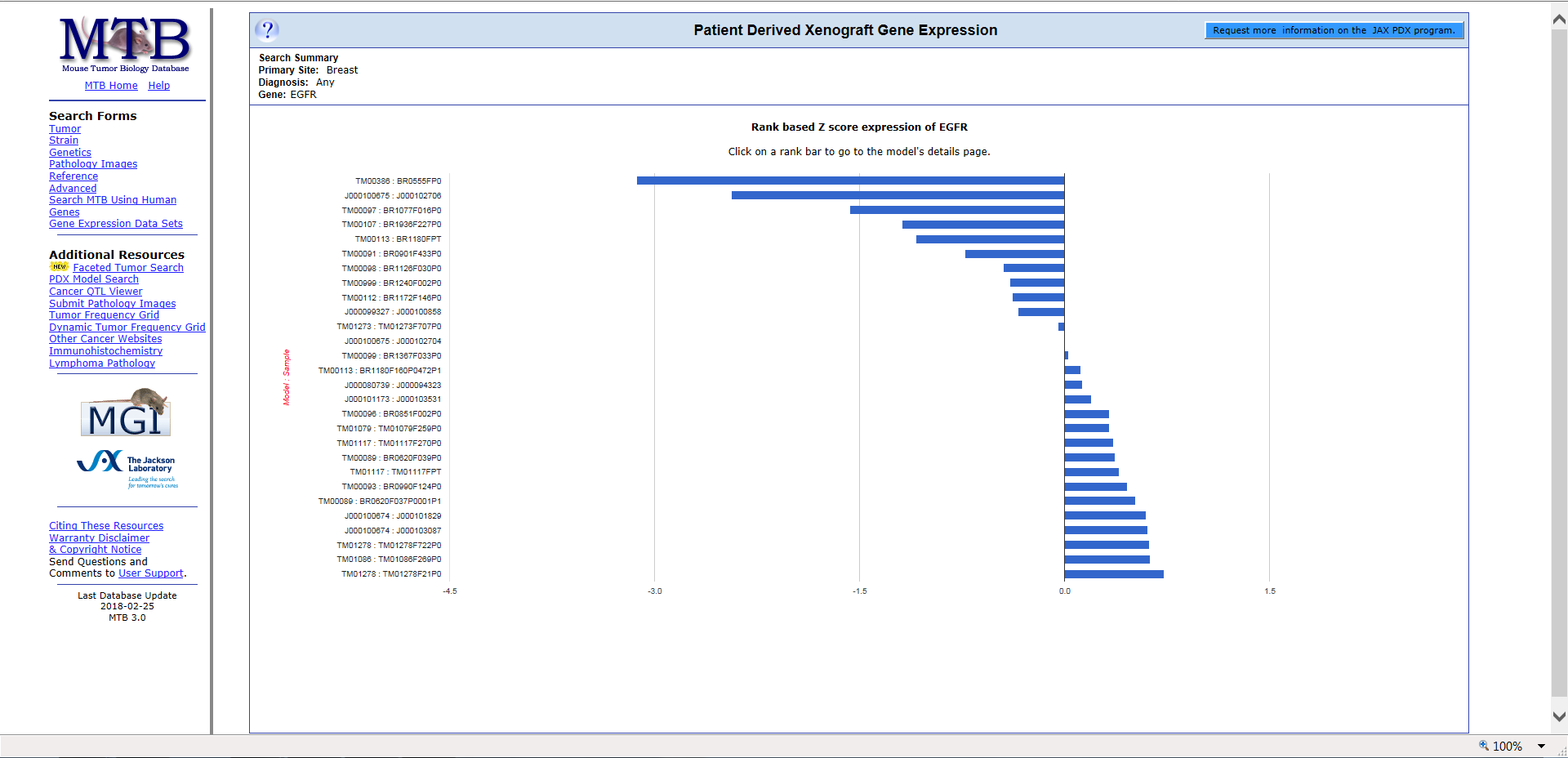
The resulting chart displays the rank-based Z score expression of EGFR, indicating where the gene expression ranks in a model compared to that gene in all models.
Be sure to check out our short video tutorial that provides a quick walkthrough of the Patient Derived Xenograft Search Form within the MTB database. Click here to view.
Next Steps
For any questions about tumor models and availability, please contact JAX Technical Information Scientists at micetech@jax.org or call I-800-422-6423 (US).
You can also confirm your tumor of interest against your target profile using formalin-fixed, paraffin-embedded (FFPE) slides even before ordering your PDX model. Traditional approaches to protein screening involve using available antibodies or molecular probes to target RNA transcription. JAX FFPE slides are available for over 400 PDX models and most can be shipped within 48 hours of ordering. These slides in combination with the MTB database enable quick and easy identification of tumors to fit your target profile.